File types¶
Authors: Enze Chen and Mark Asta (University of California, Berkeley)
Note
This is an interactive exercise, so you will want to click the and open the notebook in DataHub (or Colab for non-UCB students).
Learning objectives¶
This notebook contains exercises that explore a few of the different file types that are commonly used to store materials data. We want to give you ample hands-on practice working with different file types so that you:
Understand the advantages and disadvantages of each file format.
Are prepared to work with a variety of datasets during self-directed research.
We will progress through most of this notebook together as a group and we’re happy to answer any questions you may have about this content.
Contents¶
These exercises are grouped into the following sections:
Text files¶
From yesterday’s exercises and your own experience, you’ve worked with text files in many contexts, which speaks to their great flexibility. Indeed this is one of their strengths, as they simply consist of characters on lines that can then be represented in Python as a sequence of strings.
To revisit an example from yesterday, let’s open the file mentors.txt and print its contents.
filepath = '../../assets/data/week_1/01/mentors.txt'
with open(filepath, 'r') as f:
for line in f:
print(line.strip())
Enze
Mark
Ryan
Sinead
Pause and reflect: Why did we include the str.strip() method?
Text files are great, because we can store all sorts of information in them and they are human readable. When we talk about readability (interpretability), we typically refer to humans; but in MI, you’ll soon see that there seems to be an unfortunate tradeoff between what’s interpretable for humans and what’s interpretable for computers.
Another quick note about text files is that they don’t have to end in .txt to be a “text file.”
A Python file (.py extension) or Jupyter notebook file (.ipynb), for example, are considered text files too!
In Module 3, when you’re working with DFT calculations, you’ll likely see a lot of text files that don’t even have an extension!
(This is possible on any OS, though rarely helpful on ones with GUIs.)
When might this access pattern be less suitable?¶
We’ll go through another text file example shortly, but before we get there, we’d like to hear from you as to how you would answer this question. It seems like
with open(filepath, 'r') as f:
for line in f:
# do something
is quite powerful! But can you think of situations where we might not want to use it? 🤔
# some blank space - don't scroll too far - feel free to take notes here
A cop-out answer to the above question would be: when the file is not a text file (duh). Many such files exist, and they’re generally referred to as binary files. In fact, if you didn’t click the previous link to the Wikipedia page, just know that the first sentence literally reads: 🤣
A binary file is a computer file that is not a text file.
These are data stored as a sequence of bytes that aren’t necessarily text characters, and you’ve seen an example in yesterday’s exercises with the .npy NumPy arrays.
If we try to use the same code to read this file, we get something pretty gnarly:
with open('../../assets/data/week_1/01/2d_walk.npy', 'r') as f:
for i, line in enumerate(f):
print(line) # whoa!
if i > 2:
break # early stopping; we get the idea
“NUMPYv{'descr': '<i4', 'fortran_order': False, 'shape': (1001, 2), }
ÿÿÿÿÿÿÿÿÿÿÿÿÿÿÿÿÿÿÿÿÿÿÿÿÿÿÿÿþÿÿÿýÿÿÿýÿÿÿýÿÿÿþÿÿÿÿÿÿÿÿÿÿÿÿÿÿÿþÿÿÿýÿÿÿýÿÿÿüÿÿÿüÿÿÿýÿÿÿüÿÿÿüÿÿÿûÿÿÿûÿÿÿúÿÿÿúÿÿÿúÿÿÿúÿÿÿùÿÿÿúÿÿÿúÿÿÿùÿÿÿùÿÿÿúÿÿÿûÿÿÿüÿÿÿüÿÿÿýÿÿÿýÿÿÿýÿÿÿýÿÿÿýÿÿÿþÿÿÿþÿÿÿ þÿÿÿ ýÿÿÿýÿÿÿüÿÿÿûÿÿÿúÿÿÿûÿÿÿüÿÿÿýÿÿÿüÿÿÿüÿÿÿûÿÿÿûÿÿÿûÿÿÿüÿÿÿüÿÿÿýÿÿÿýÿÿÿüÿÿÿüÿÿÿüÿÿÿûÿÿÿüÿÿÿûÿÿÿûÿÿÿûÿÿÿúÿÿÿùÿÿÿøÿÿÿ÷ÿÿÿ÷ÿÿÿøÿÿÿøÿÿÿùÿÿÿúÿÿÿûÿÿÿúÿÿÿùÿÿÿøÿÿÿøÿÿÿøÿÿÿøÿÿÿøÿÿÿøÿÿÿøÿÿÿùÿÿÿùÿÿÿùÿÿÿøÿÿÿ÷ÿÿÿöÿÿÿöÿÿÿõÿÿÿõÿÿÿõÿÿÿôÿÿÿóÿÿÿóÿÿÿóÿÿÿóÿÿÿòÿÿÿòÿÿÿóÿÿÿóÿÿÿóÿÿÿóÿÿÿôÿÿÿôÿÿÿôÿÿÿôÿÿÿõÿÿÿõÿÿÿöÿÿÿ÷ÿÿÿöÿÿÿõÿÿÿõÿÿÿöÿÿÿõÿÿÿõÿÿÿôÿÿÿôÿÿÿóÿÿÿóÿÿÿóÿÿÿôÿÿÿõÿÿÿöÿÿÿõÿÿÿõÿÿÿõÿÿÿöÿÿÿöÿÿÿöÿÿÿõÿÿÿöÿÿÿöÿÿÿõÿÿÿöÿÿÿ÷ÿÿÿ÷ÿÿÿ÷ÿÿÿøÿÿÿùÿÿÿùÿÿÿùÿÿÿùÿÿÿùÿÿÿúÿÿÿûÿÿÿúÿÿÿúÿÿÿúÿÿÿûÿÿÿüÿÿÿýÿÿÿüÿÿÿüÿÿÿüÿÿÿüÿÿÿüÿÿÿýÿÿÿýÿÿÿýÿÿÿþÿÿÿþÿÿÿþÿÿÿþÿÿÿþÿÿÿþÿÿÿþÿÿÿÿÿÿÿÿÿÿÿÿÿÿÿÿÿÿÿÿÿÿÿÿÿÿÿþÿÿÿÿÿÿÿÿÿÿÿÿÿÿÿþÿÿÿýÿÿÿüÿÿÿüÿÿÿüÿÿÿûÿÿÿúÿÿÿúÿÿÿùÿÿÿúÿÿÿûÿÿÿúÿÿÿúÿÿÿûÿÿÿûÿÿÿúÿÿÿúÿÿÿûÿÿÿúÿÿÿúÿÿÿúÿÿÿûÿÿÿûÿÿÿüÿÿÿüÿÿÿýÿÿÿþÿÿÿýÿÿÿüÿÿÿûÿÿÿûÿÿÿüÿÿÿüÿÿÿüÿÿÿûÿÿÿüÿÿÿüÿÿÿûÿÿÿúÿÿÿûÿÿÿûÿÿÿûÿÿÿúÿÿÿúÿÿÿúÿÿÿùÿÿÿøÿÿÿøÿÿÿøÿÿÿøÿÿÿùÿÿÿøÿÿÿøÿÿÿøÿÿÿøÿÿÿøÿÿÿøÿÿÿ÷ÿÿÿ÷ÿÿÿ÷ÿÿÿøÿÿÿùÿÿÿùÿÿÿùÿÿÿùÿÿÿøÿÿÿøÿÿÿøÿÿÿøÿÿÿøÿÿÿøÿÿÿøÿÿÿøÿÿÿùÿÿÿùÿÿÿøÿÿÿÿÿÿÿøÿÿÿøÿÿÿÿÿÿÿøÿÿÿøÿÿÿÿÿÿÿøÿÿÿøÿÿÿÿÿÿÿøÿÿÿøÿÿÿ÷ÿÿÿøÿÿÿùÿÿÿøÿÿÿùÿÿÿøÿÿÿ÷ÿÿÿÿÿÿÿ÷ÿÿÿ÷ÿÿÿÿÿÿÿ÷ÿÿÿ÷ÿÿÿÿÿÿÿ÷ÿÿÿþÿÿÿ÷ÿÿÿýÿÿÿ÷ÿÿÿýÿÿÿøÿÿÿþÿÿÿøÿÿÿþÿÿÿùÿÿÿýÿÿÿùÿÿÿýÿÿÿøÿÿÿýÿÿÿ÷ÿÿÿýÿÿÿöÿÿÿüÿÿÿöÿÿÿüÿÿÿ÷ÿÿÿüÿÿÿöÿÿÿüÿÿÿ÷ÿÿÿýÿÿÿ÷ÿÿÿýÿÿÿøÿÿÿüÿÿÿøÿÿÿüÿÿÿùÿÿÿýÿÿÿùÿÿÿýÿÿÿøÿÿÿüÿÿÿøÿÿÿüÿÿÿ÷ÿÿÿûÿÿÿ÷ÿÿÿúÿÿÿ÷ÿÿÿúÿÿÿöÿÿÿûÿÿÿöÿÿÿûÿÿÿõÿÿÿûÿÿÿôÿÿÿúÿÿÿôÿÿÿúÿÿÿõÿÿÿúÿÿÿöÿÿÿûÿÿÿöÿÿÿûÿÿÿõÿÿÿûÿÿÿöÿÿÿûÿÿÿõÿÿÿúÿÿÿõÿÿÿùÿÿÿõÿÿÿøÿÿÿõÿÿÿøÿÿÿôÿÿÿøÿÿÿõÿÿÿùÿÿÿõÿÿÿøÿÿÿõÿÿÿøÿÿÿöÿÿÿøÿÿÿ÷ÿÿÿùÿÿÿ÷ÿÿÿùÿÿÿøÿÿÿùÿÿÿ÷ÿÿÿøÿÿÿ÷ÿÿÿøÿÿÿøÿÿÿùÿÿÿøÿÿÿùÿÿÿùÿÿÿùÿÿÿøÿÿÿøÿÿÿøÿÿÿøÿÿÿùÿÿÿ÷ÿÿÿùÿÿÿöÿÿÿùÿÿÿ÷ÿÿÿùÿÿÿ÷ÿÿÿúÿÿÿ÷ÿÿÿûÿÿÿöÿÿÿûÿÿÿ÷ÿÿÿûÿÿÿ÷ÿÿÿúÿÿÿ÷ÿÿÿûÿÿÿöÿÿÿûÿÿÿöÿÿÿüÿÿÿöÿÿÿýÿÿÿöÿÿÿüÿÿÿõÿÿÿüÿÿÿõÿÿÿýÿÿÿôÿÿÿýÿÿÿôÿÿÿþÿÿÿôÿÿÿÿÿÿÿôÿÿÿôÿÿÿÿÿÿÿôÿÿÿõÿÿÿöÿÿÿõÿÿÿõÿÿÿõÿÿÿôÿÿÿóÿÿÿóÿÿÿÿÿÿÿóÿÿÿþÿÿÿóÿÿÿÿÿÿÿóÿÿÿóÿÿÿòÿÿÿóÿÿÿóÿÿÿóÿÿÿÿÿÿÿóÿÿÿóÿÿÿóÿÿÿóÿÿÿÿÿÿÿôÿÿÿÿÿÿÿôÿÿÿþÿÿÿóÿÿÿþÿÿÿòÿÿÿþÿÿÿòÿÿÿýÿÿÿñÿÿÿýÿÿÿòÿÿÿýÿÿÿñÿÿÿýÿÿÿñÿÿÿüÿÿÿñÿÿÿûÿÿÿñÿÿÿüÿÿÿñÿÿÿýÿÿÿñÿÿÿþÿÿÿðÿÿÿþÿÿÿñÿÿÿþÿÿÿñÿÿÿÿÿÿÿðÿÿÿÿÿÿÿïÿÿÿÿÿÿÿîÿÿÿÿÿÿÿîÿÿÿþÿÿÿîÿÿÿÿÿÿÿïÿÿÿÿÿÿÿðÿÿÿÿÿÿÿðÿÿÿðÿÿÿðÿÿÿðÿÿÿðÿÿÿðÿÿÿðÿÿÿðÿÿÿðÿÿÿñÿÿÿñÿÿÿòÿÿÿòÿÿÿñÿÿÿñÿÿÿñÿÿÿñÿÿÿÿÿÿÿñÿÿÿñÿÿÿðÿÿÿïÿÿÿîÿÿÿïÿÿÿðÿÿÿïÿÿÿïÿÿÿïÿÿÿïÿÿÿðÿÿÿðÿÿÿñÿÿÿòÿÿÿñÿÿÿñÿÿÿðÿÿÿñÿÿÿðÿÿÿñÿÿÿðÿÿÿðÿÿÿñÿÿÿñÿÿÿñÿÿÿðÿÿÿðÿÿÿðÿÿÿðÿÿÿðÿÿÿðÿÿÿñÿÿÿñÿÿÿòÿÿÿñÿÿÿñÿÿÿðÿÿÿïÿÿÿïÿÿÿïÿÿÿðÿÿÿïÿÿÿîÿÿÿîÿÿÿîÿÿÿîÿÿÿîÿÿÿïÿÿÿïÿÿÿÿÿÿÿïÿÿÿþÿÿÿîÿÿÿþÿÿÿîÿÿÿÿÿÿÿîÿÿÿîÿÿÿÿÿÿÿïÿÿÿÿÿÿÿïÿÿÿïÿÿÿïÿÿÿïÿÿÿîÿÿÿîÿÿÿïÿÿÿðÿÿÿðÿÿÿÿÿÿÿðÿÿÿïÿÿÿïÿÿÿïÿÿÿîÿÿÿîÿÿÿíÿÿÿîÿÿÿîÿÿÿîÿÿÿïÿÿÿðÿÿÿðÿÿÿñÿÿÿòÿÿÿòÿÿÿñÿÿÿñÿÿÿñÿÿÿòÿÿÿñÿÿÿòÿÿÿòÿÿÿñÿÿÿñÿÿÿñÿÿÿðÿÿÿñÿÿÿñÿÿÿñÿÿÿñÿÿÿñÿÿÿñÿÿÿÿÿÿÿðÿÿÿÿÿÿÿðÿÿÿþÿÿÿðÿÿÿÿÿÿÿðÿÿÿïÿÿÿïÿÿÿÿÿÿÿîÿÿÿÿÿÿÿîÿÿÿþÿÿÿíÿÿÿþÿÿÿìÿÿÿþÿÿÿëÿÿÿþÿÿÿêÿÿÿþÿÿÿêÿÿÿÿÿÿÿéÿÿÿÿÿÿÿêÿÿÿÿÿÿÿêÿÿÿþÿÿÿêÿÿÿýÿÿÿëÿÿÿýÿÿÿëÿÿÿþÿÿÿìÿÿÿþÿÿÿëÿÿÿþÿÿÿëÿÿÿÿÿÿÿëÿÿÿìÿÿÿíÿÿÿîÿÿÿïÿÿÿîÿÿÿïÿÿÿïÿÿÿïÿÿÿïÿÿÿðÿÿÿñÿÿÿðÿÿÿðÿÿÿñÿÿÿòÿÿÿóÿÿÿóÿÿÿóÿÿÿóÿÿÿóÿÿÿóÿÿÿòÿÿÿóÿÿÿôÿÿÿóÿÿÿòÿÿÿòÿÿÿñÿÿÿñÿÿÿñÿÿÿñÿÿÿðÿÿÿðÿÿÿïÿÿÿïÿÿÿîÿÿÿíÿÿÿîÿÿÿïÿÿÿïÿÿÿïÿÿÿðÿÿÿðÿÿÿðÿÿÿðÿÿÿñÿÿÿñÿÿÿñÿÿÿðÿÿÿðÿÿÿïÿÿÿðÿÿÿñÿÿÿñÿÿÿñÿÿÿðÿÿÿñÿÿÿðÿÿÿðÿÿÿðÿÿÿïÿÿÿïÿÿÿîÿÿÿîÿÿÿïÿÿÿîÿÿÿïÿÿÿïÿÿÿðÿÿÿñÿÿÿðÿÿÿðÿÿÿïÿÿÿïÿÿÿïÿÿÿïÿÿÿðÿÿÿïÿÿÿðÿÿÿñÿÿÿòÿÿÿòÿÿÿñÿÿÿðÿÿÿðÿÿÿïÿÿÿïÿÿÿïÿÿÿïÿÿÿ ïÿÿÿ
ðÿÿÿ
ïÿÿÿ
When the file is structured¶
A more subtle answer to the above question would be when the file is still a text file, but the text is written in a way that has some internal structure. The flexibility of text files is really nice if we’re writing prose or poetry, but data are usually more systematically organized to facilitate access. This means there might be a more specialized/targeted way to read in the contents of the file than going line by line and interpreting the contents as strings, particularly as a lot of data are numeric. As we move forward and discuss different examples of structured data, we invite you to think about what we said earlier about the tradeoff between human interpretability and computer interpretability.
Exercise: more elemental properties¶
For this next exercise, we’ll attempt to do some live coding as a group. This exercise will give us practice working with files, control flow, lists, and casting.
Inside the text file hardness_density.csv (CSV is another type of text file), we have some data organized as follows:
# Data obtained from Wikipedia
Element,Number,Mohs hardness,Density (g/cc)
lithium,3,0.6,0.534
beryllium,4,5.5,1.85
boron,5,9.4,2.34
carbon,6,10,3.513
...
Our job is to read in this text file and store the data, starting from the third line, as a list of lists.
The outer list has an element for each line, and the inner list should have elements of type [str, int, float, float] representing the element, atomic number, Mohs hardness, and density, respectively.
data =
[['lithium', 3, 0.6, 0.534],
['beryllium', 4, 5.5, 1.85],
['boron', 5, 9.4, 2.34],
['carbon', 6, 10.0, 3.513],
...
]
filepath = '../../assets/data/week_1/02/hardness_density.csv'
# ------------- WRITE YOUR CODE IN THE SPACE BELOW ---------- #
Pause and reflect: Is there any structure in the above data? We will revisit this example in the next lesson!
JSON files¶
A CSV file is an example of a text file with some structure, but there’s so much to say on this topic that we’ll dedicate an entire lesson to it. In the meantime, we’ll explore another kind of structured text file that is very commonly used for materials data, and that is JavaScript Object Notation, or JSON for short. It is very commonly used to store hierarchical data in array structures (like lists!) and key-value pairs (like dictionaries!), both of which we’re familiar with, so it makes sense to discuss this next. An example of what a JSON entry for the above data would look like could be:
[
{
"element": "lithium",
"number": 3,
"hardness": 0.6,
"density": {
"value": 0.534,
"units": "g/cc",
}
},
{
"element": "beryllium",
...
}
]
Pause and reflect: What data types and data structures do we see in the snippet above?
Fundamentally, a JSON file is just another text file.
For example, we have some band gap data from the canonical dataset compiled by Strehlow and Cook, J. Phys. Chem. Ref. Data, 1973 stored in the file band_gaps_sc.json, which was obtained from Citrination.
We could, if we want to, simply try to read this file from top to bottom like so:
filepath = '../../assets/data/week_1/02/band_gaps_sc.json'
with open(filepath, 'r') as f:
for i, line in enumerate(f):
print(line.rstrip()) # this removes invisibles from the right side of the string ONLY
if i > 25:
break # early stopping, we get the idea
[
{
"category": "system.chemical",
"references": [
{
"doi": "10.1063/1.3253115"
}
],
"properties": [
{
"name": "Crystallinity",
"scalars": [
{
"value": "Single crystalline"
}
]
},
{
"name": "Band gap",
"scalars": "13.6",
"units": "eV",
"conditions": [
{
"name": "Transition",
"scalars": [
{
"value": "Direct"
While this works, it feels clunky if we wanted to extract all band gap values and we wouldn’t be able to take advantage of all the nice indexing features that come with lists and dictionaries. If only there was a way for us to load in the entire file as a list of dictionaries representing our materials…
json to the rescue¶

Fortunately, there exists another package in Python that allows us to do that, and this package is called, appropriately, json.
We first import the json package and then use it as follows:
import json
filepath = '../../assets/data/week_1/02/band_gaps_sc.json'
with open(filepath, 'r') as f: # same as any other text file
materials = json.load(f) # special function from the json package!
# display the first material in the dataset
print(f"'materials' is a variable of type {type(materials)}.")
print(f'Its elements are of type {type(materials[0])}.')
print(f'There are {len(materials)} materials in total. Not that many!')
print('The first element is the following:')
materials[0]
'materials' is a variable of type <class 'list'>.
Its elements are of type <class 'dict'>.
There are 1447 materials in total. Not that many!
The first element is the following:
{'category': 'system.chemical',
'references': [{'doi': '10.1063/1.3253115'}],
'properties': [{'name': 'Crystallinity',
'scalars': [{'value': 'Single crystalline'}]},
{'name': 'Band gap',
'scalars': '13.6',
'units': 'eV',
'conditions': [{'name': 'Transition', 'scalars': [{'value': 'Direct'}]},
{'name': 'Temperature', 'scalars': [{'value': '300'}], 'units': 'K'}],
'method': {'name': 'Reflection'},
'dataType': 'EXPERIMENTAL'}],
'chemicalFormula': 'Li1F1'}
Aha! Now our data are represented by sensible Python constructs. In particular, you’ll notice that the JSON allows metadata to be intuitively associated with the corresponding data (e.g., the temperature at which a particular measurement was made). Let’s see if we can dig a little deeper and extract some useful data out of this dataset.
Exercise (small group): create a list that stores all of the band gap values in the Strehlow and Cook dataset¶
Your first few values should be: band_gaps = [13.6, 12.61, 12.6, 12.1, 12, ...]
Hints:
Band gap is stored in the
propertiesentry of the dictionary, but not every material has every property, so the list (value) associated with this key will have varying lengths!How can we ensure we’ve found the
"Band gap"property?You may assume that every material has a band gap associated with it.
band_gaps = []
# ------------- WRITE YOUR CODE IN THE SPACE BELOW ---------- #
# --------------------------------------------------------------- #
band_gaps[:10]
[]
Writing JSON files¶
If you have some hierarchical data stored inside lists and dictionaries, you may want to save it to an external JSON file.
As a very simple example, let’s take the list of band gap values you created in the previous exercise and try writing that list (band_gaps) to a JSON (for any other data, the code is the same, and only the file content structure/hierarchy will be different).
We use the json.dump() function as follows:
with open('path/to/output_file.json', 'w') as f: # note the 'w' for write!
json.dump(data_object, f, indent=4) # indent=4 is a best practice
When applied to our data, the result is:
with open('../../assets/data/week_1/02/band_gaps_only.json', 'w') as f:
json.dump(band_gaps, f, indent=4)
Versatility¶
JSON files are very good at storing hierarchical data, which makes them suitable for many materials data, as explained by Kyle and Bryce in their paper. But JSON is used in many other applications too, such as this very notebook that you’re currently reading! 😉
Images?¶
While we will not have time to teach you about image data in this module, we will briefly show how they can be handled in Python. Given the importance and diversity of materials characterization techniques, it would be remiss of us to not at least mention image analysis. You have probably heard a lot about this topic in the media in the context of self-driving cars, deep fakes, and medicine just to name a few, and it’s becoming a big deal in MI too.
There are many packages out there for working with image data, so for the purposes of demonstration, we will work with the standard Python Imaging Library (PIL), now developed under the Pillow project, for reading an image file.
# Import modules
from PIL import Image
img = Image.open('../../assets/fig/week_1/02/microstructure.png')
# img = Image.open('../../assets/fig/week_1/02/diffraction.jpg')
# We can even convert the image into a NumPy array
import numpy as np
img_arr = np.asarray(img)
print(f'The image has dimensions {img_arr.shape}.') # what are all the dimensions?
# Display the PIL Image
img
The image has dimensions (768, 960, 3).
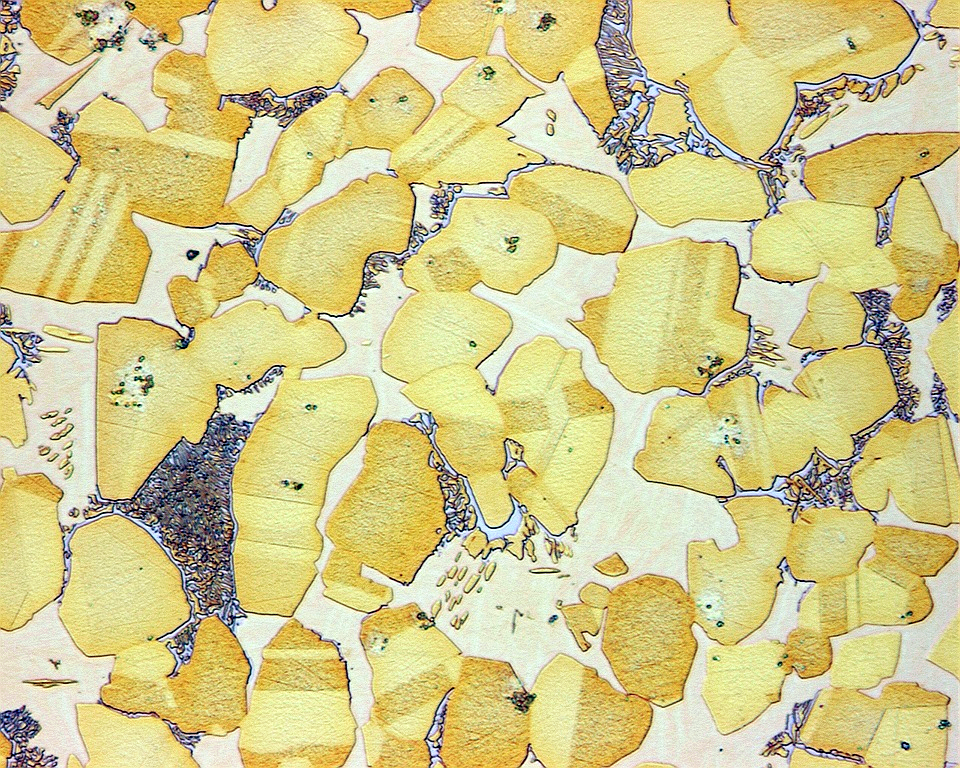
While this is all we have time for today, fear not, because Alex will be speaking to us next Wednesday about her own research at the intersection of MI and electron microscopy! For more information on data-driven electron microscopy, you can also check out this commentary in Nature Materials by the leading researchers in this field.
When files simply aren’t enough¶
At this point, you’ve seen a lot of different files types for storing materials data. Specifically, we’ve discussed regular text files, binary files, CSVs, JSONs, and images, and you’ll get more practice working with several of these in this module. This is already an incredibly diverse set of data, which speaks to a lot of the challenges faced in MI.
However, there will be times when our data get so large and complex that it becomes infeasible to store them in files, even if these files are stored in the cloud. This calls for something else—this calls for databases! Stay tuned for more. 🙂
Conclusion¶
This concludes our discussion of files. 📄📂 Up next, we’re going to take a deep dive into tabular data and CSV files specifically. Please don’t hesitate to reach out on Slack if you have questions or concerns about this content.