Introduction to tabular data in Python
Contents
Introduction to tabular data in Python¶
Authors: Enze Chen (University of California, Berkeley)
Note
This is an interactive exercise, so you will want to click the and open the notebook in DataHub (or Colab for non-UCB students).
This notebook contains a series of exercises that explore tabular data and the pandas package. We hope that after going through these exercises, you will be prepared to use pandas and other programmatic tools to operate on tabular data, which can be very helpful when analyzing X-ray diffraction (XRD) data. Like the previous lab, if you’re already familiar working with tabular data, you can jump straight to the Lab 2 exercises on the next page.
Contents¶
These exercises are grouped into the following sections:
File formats¶
We’ll begin by discussing a common problem faced by materials scientists: working with different file types. Particularly in industry, and particularly when working with collaborators, you’ll be receiving information in different formats including:
Text-based files where the information is completely unstructured.
Text-based files with hierarchical structure, such as JSON.
Image files with all sorts of file extensions and compression formats.
PDFs which may or may not be formatted for optical character recognition (OCR).
Binary files with machine-readable data.
Proprietary data formats that can only be opened with special software!
The lack of standardization is definitely a pain and a bottleneck in data science pipelines when working with materials data. Therefore, we’re using this lesson as an opportunity to learn how to better interface with data files programmatically.
CSV files¶
By now, you’ve probably realized that your XRD data is highly structured.
In particular, it takes the format angle,intensity inside a text file, like so:
3.00,1712.5
3.02,1850
3.04,2075
3.06,1912.5
...
We call these files comma-separated values files, or CSV for short. They’re nice because they’re easy to store and they strike a healthy balance between easy for humans to read (looks like a data table) and easy for computers to read (the values can be loaded into an array). Previously, you’ve seen how the NumPy package can read in CSV files for plotting purposes, but we didn’t say much more. Here, we’ll teach you how to use another package that gives you even more flexibility when working with such tabular data.
pandas!¶

pandas is Python package developed in 2008 to facilitate data analysis and it has since become one of the most widely used packages in the Python community.
The name is derived from “panel data” and the package is built on top of NumPy, which you should already be familiar with!
DataFrames¶
At the heart of pandas lies the DataFrame data structure for all of your informatics needs.
The DataFrame follows our conventional notion of a spreadsheet for 2D tabular data.
Note that a DataFrame is for 2D data only and does not work for lower or higher dimensions (unlike NumPy arrays).
They feature row and column labels for intelligent indexing and different columns can contain different types (a mix of int, str, bool, etc.) 🙌
To import pandas, the community standard is to use the alias
import pandas as pd
There are many ways to create DataFrames, but with the DataFrame() constructor you will likely use one of two methods:
pd.DataFrame(dict), wheredictis a dictionary ofstr:listpairs that correspond to the name and values in each column.pd.DataFrame(arr, columns=col_names), wherearris a 2D NumPy array or list of lists andcol_namesare the names you want to assign each column.
You’ll likely find yourself using both methods when building DataFrames from scratch, all depends on your use case. Let’s see this in practice.
import pandas as pd
# Constructor method 1
number = [1, 2, 3]
mass = [1, 4, 7]
df1 = pd.DataFrame({'atomic number':number, 'atomic mass':mass}) # df is common shorthand for DataFrame
display(df1)
# Constructor method 2
arr = [[1, 1], [2, 4], [3, 7]]
df2 = pd.DataFrame(arr, columns=['atomic number', 'atomic mass'])
display(df2)
| atomic number | atomic mass | |
|---|---|---|
| 0 | 1 | 1 |
| 1 | 2 | 4 |
| 2 | 3 | 7 |
| atomic number | atomic mass | |
|---|---|---|
| 0 | 1 | 1 |
| 1 | 2 | 4 |
| 2 | 3 | 7 |
And the nice part about working with pandas DataFrames in Jupyter notebooks is the stylistic rendering! 🙏🏼 Take a moment to see how the column names in the constructors correspond to the names in the DataFrame; in particular, note that these names are labels in the DataFrame, but they’re not part of the data itself.
Reading in a CSV file¶
What if your data source is a CSV file that’s already nicely formatted for you? It would be nice if we could directly use the structure in our data and express it in Python—you don’t want to be stuck typing out all those rows.
To do this, we will use the pd.read_csv() function which does exactly as the name describes.
This function, as you can see from the documentation, has a bajillion input arguments that we don’t have time to cover today, so we’ll describe just a few here:
filepath_or_buffer(str): The only required argument, which is the file you’re trying to read in.sep(str): The character separating your data tokens. Default is a comma (','), but you can change this to be space (' '), tab ('\t'), etc.header(int): Which row to read column headers from. Default is the first row.names(list): If you would like to rename your columns. This list cannot have duplicates.skiprows(int): Extremely useful if there are extra lines at the top of the file that you want to avoid reading (e.g., comments).nrows(int): Extremely useful if your file is very large or if your data stops before reaching the end of the file (e.g., there’s a footer).
Notice how this is similar to np.loadtxt(), just with a different default separator and some extra headers for the columns (bolded in the previous output).
We’ll address these headers shortly.
Let’s use pd.read_csv() to read in our sine.txt file from Lab 1’s demo (copied again into this folder).
We’ll also rename the column names to be:
['x', 'sin']
df_sine = pd.read_csv('sine.txt', names=['x', 'sin']) # try omitting the names argument and see what happens
df_sine
| x | sin | |
|---|---|---|
| 0 | 0.000000 | 0.000000 |
| 1 | 0.006283 | 0.006283 |
| 2 | 0.012566 | 0.012566 |
| 3 | 0.018850 | 0.018848 |
| 4 | 0.025133 | 0.025130 |
| ... | ... | ... |
| 996 | 6.258053 | -0.025130 |
| 997 | 6.264336 | -0.018848 |
| 998 | 6.270619 | -0.012566 |
| 999 | 6.276902 | -0.006283 |
| 1000 | 6.283185 | -0.000000 |
1001 rows × 2 columns
Working with DataFrames¶
We got our data into a DataFrame! Now we can have some fun. 😄
The first thing you might be interested in is the dimensions of your data (DataFrame).
We can access that using the df.shape attribute, which returns (rows, columns).
This is often a helpful sanity check.
df_sine.shape
(1001, 2)
We can also get statistics of the rows/columns of the DataFrame by using methods like df.min(), df.max(), and df.mean().
For these methods, the axis parameter specifies if we compute along the columns (axis=0) or rows (axis=1).
df_sine.min() # default is axis=0, compute along columns.
# note these minima are in each INDIVIDUAL column,
# NOT necessarily corresponding to the same data point (row)!
x 0.0
sin -1.0
dtype: float64
df_sine.mean(axis=1) # compute mean along each row
0 0.000000
1 0.006283
2 0.012566
3 0.018849
4 0.025131
...
996 3.116461
997 3.122744
998 3.129026
999 3.135309
1000 3.141593
Length: 1001, dtype: float64
Indexing and slicing a DataFrame¶
There are many ways to index into a DataFrame and slice it to get subsets of your data. If we want to get the data in a single column, all we have to do is write the column name as a string! For example:
df_sine['x'] # this gets the x column of the DataFrame
0 0.000000
1 0.006283
2 0.012566
3 0.018850
4 0.025133
...
996 6.258053
997 6.264336
998 6.270619
999 6.276902
1000 6.283185
Name: x, Length: 1001, dtype: float64
Combining columns with operations¶
Recall that with NumPy arrays, we could perform arithmetic operations on the entire array or a slice of it and store the result somewhere. The neat thing about pandas DataFrames is that if we apply an operation to an entire column, we can store the result in a new column that’s appended to the original DataFrame! The general syntax is:
df['new_column_name'] = operation(df['old_column_name'])
See the example below where we use np.cos() to compute the cosine of the x values in our previous DataFrame.
import numpy as np
df_sine['cos'] = np.cos(df_sine['x'])
df_sine # ta-da!
| x | sin | cos | |
|---|---|---|---|
| 0 | 0.000000 | 0.000000 | 1.000000 |
| 1 | 0.006283 | 0.006283 | 0.999980 |
| 2 | 0.012566 | 0.012566 | 0.999921 |
| 3 | 0.018850 | 0.018848 | 0.999822 |
| 4 | 0.025133 | 0.025130 | 0.999684 |
| ... | ... | ... | ... |
| 996 | 6.258053 | -0.025130 | 0.999684 |
| 997 | 6.264336 | -0.018848 | 0.999822 |
| 998 | 6.270619 | -0.012566 | 0.999921 |
| 999 | 6.276902 | -0.006283 | 0.999980 |
| 1000 | 6.283185 | -0.000000 | 1.000000 |
1001 rows × 3 columns
Here’s another example performing subtraction of one column by another:
df_sine['diff (x - sin)'] = df_sine['x'] - df_sine['sin']
df_sine
| x | sin | cos | diff (x - sin) | |
|---|---|---|---|---|
| 0 | 0.000000 | 0.000000 | 1.000000 | 0.000000e+00 |
| 1 | 0.006283 | 0.006283 | 0.999980 | 4.100000e-08 |
| 2 | 0.012566 | 0.012566 | 0.999921 | 3.310000e-07 |
| 3 | 0.018850 | 0.018848 | 0.999822 | 1.116000e-06 |
| 4 | 0.025133 | 0.025130 | 0.999684 | 2.646000e-06 |
| ... | ... | ... | ... | ... |
| 996 | 6.258053 | -0.025130 | 0.999684 | 6.283183e+00 |
| 997 | 6.264336 | -0.018848 | 0.999822 | 6.283184e+00 |
| 998 | 6.270619 | -0.012566 | 0.999921 | 6.283185e+00 |
| 999 | 6.276902 | -0.006283 | 0.999980 | 6.283185e+00 |
| 1000 | 6.283185 | -0.000000 | 1.000000 | 6.283185e+00 |
1001 rows × 4 columns
Here’s yet another example where we simply tack on a new column of values.
df_sine['foo'] = np.arange(1, df_sine.shape[0] + 1)
df_sine
| x | sin | cos | diff (x - sin) | foo | |
|---|---|---|---|---|---|
| 0 | 0.000000 | 0.000000 | 1.000000 | 0.000000e+00 | 1 |
| 1 | 0.006283 | 0.006283 | 0.999980 | 4.100000e-08 | 2 |
| 2 | 0.012566 | 0.012566 | 0.999921 | 3.310000e-07 | 3 |
| 3 | 0.018850 | 0.018848 | 0.999822 | 1.116000e-06 | 4 |
| 4 | 0.025133 | 0.025130 | 0.999684 | 2.646000e-06 | 5 |
| ... | ... | ... | ... | ... | ... |
| 996 | 6.258053 | -0.025130 | 0.999684 | 6.283183e+00 | 997 |
| 997 | 6.264336 | -0.018848 | 0.999822 | 6.283184e+00 | 998 |
| 998 | 6.270619 | -0.012566 | 0.999921 | 6.283185e+00 | 999 |
| 999 | 6.276902 | -0.006283 | 0.999980 | 6.283185e+00 | 1000 |
| 1000 | 6.283185 | -0.000000 | 1.000000 | 6.283185e+00 | 1001 |
1001 rows × 5 columns
Quick exercise: Add another column to the DataFrame to verify the identity¶
Recall that exponentiation is base ** power.
# ------------- WRITE YOUR CODE IN THE SPACE BELOW ---------- #
Plotting data from a DataFrame¶
We can easily index into a DataFrame using the headers to select the columns for plotting!
import matplotlib.pyplot as plt
fig, ax = plt.subplots()
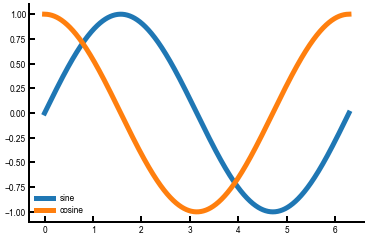
ax.plot(df_sine['x'], df_sine['sin'], label='sine')
ax.plot(df_sine['x'], df_sine['cos'], label='cosine')
ax.legend()
plt.show()
Saving a DataFrame¶
There are many ways to save the data, in text, binary, etc., but the easiest way for purely numerical data is probably as a text (CSV) file using df.to_csv(filename).
Some things to be aware of:
pandas likes to save the index (row labels) by default, but that looks a little weird if you later open the CSV in a program like Excel, so we typically like to have
index=Falseas another input argument.Note that this is a method attached to a DataFrame object, not the pandas package! That is, it’s
df.to_csv(), notpd.to_csv(df).
After you run the following cell, you should see the new TXT file appear in your JupyterHub folder.
df_sine.to_csv('sine_updated.txt', index=False) # save the df we've been working with
Conclusion¶
This concludes the introduction to tabular data. Realistically, there’s way too much to cover about pandas, and a lot of your understanding will be built as you work with it for your own datasets (official user guide). But, we hope that by giving a brief introduction, you at least know what’s available, where to go for help, and how to think about tabular data with pandas, at least enough to finish up the lab calculations. Onwards to the exercises!